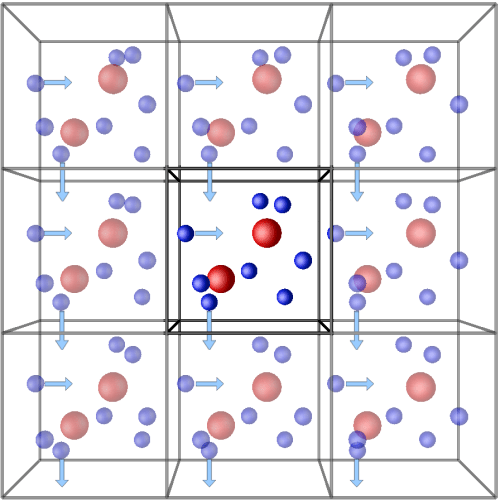
Our system of interest, whatever it is, in a simulation is a smaller part of something bigger. However, things behave differently in a “more natural” environment versus surrounded by nothing–vacuum–on all sides. We don’t want these surface effects, so we try to simulate our system in a more natural setting by surrounding it (the unit cell) with a number of copies of itself (images). Then, if a particle leaves the original simulation box, an identical image particle enters the original box from the opposite side. The number of particles in the original box therefore stays constant. These conditions are known as periodic boundary conditions.
How big do we want our original box’s unit cell?
- Big enough such that the solute (protein or whatever) can’t see or interact with itself in the image boxes.
- Small enough such that we’re not wasting resources simulating a bunch of excess solvent far from the solute.
What shape is the box?
It’s easiest to think of just a standard cubic cell, which is replicated on all front/back/left/right/top/bottom sides. Different shapes are possible, and may be preferred for specific simulations. Whatever shape is chosen has to be able to translate and fill the space without gaps in between cells. Options boil down to:
Example: The truncated octahedron unit cell may be more appropriate for spherical solutes. The distance between neighboring cells is greater than for a conventional cube, so fewer particles can be used in the cell.
According to the GROMACS manual (p. 13),
The rhombic dodecahedron is the smallest and most regular space-filling unit cell. Each of the 12 image cells is at the same distance. The volume is 71% of the volume of a cube having the same image distance. This saves about 29% of CPU-time when simulating a spherical or flexible molecule in solvent.
Which particles interact with which?
A common form of handling particle interactions is using the minimum image convention, in which a particle sees only the closest image of all the other particles in the simulation. In other words, we don’t want to double count the solute interacting with the same specific water molecule twice, for example.
Additional resources
- Molecular Modelling: Principles and Applications. Leach. pp. 317-319, 324.